Protein & Antibody Engineering
Building for Biology
Engineering a Co-Stimulatory T-cell Agonist
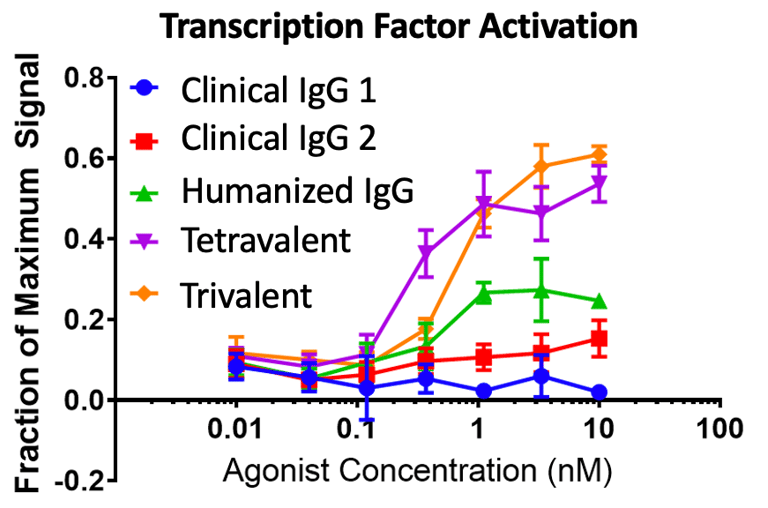
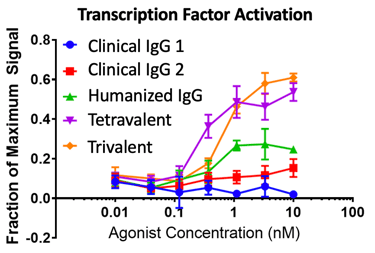
One great project was developing a soluble T-cell co-stimulatory agonist. We used two CROs to isolate leads: one immunized mice with native ligand and the other used a display platform/synthetic library. The murine campaign yielded ~90 high-affinity (10-100pM) antibodies against the same epitope, and the synthetic library campaign yielded ~60 antibodies of moderate affinity (50-100nM) against four different epitopes. The epitope mattered here, and we were lucky to get good leads from the immunization campaign. The leads competed with the native ligand and one previously established clinical antibody (not shown). We humanized the leads and had a fun time making some great higher valency molecules, and we found that our tri- and tetravalent formats did much better in a transgenic receptor/downstream transcription factor activation assay (below) than the clinical antibodies. The hi-valency agonists also induced T-cells to make as much cytokine as the clinical antibodies and rescued the cells from anergy in our assays.
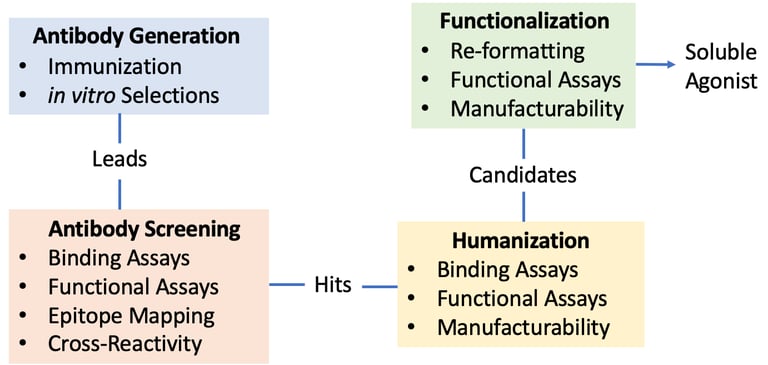
Engineering a B-cell Agonist
Confronted with the high-costs of reagents for B-cell manufacturing, The team engineered and tested a soluble B-cell agonist in a project similar to the T-cell agonist project above.
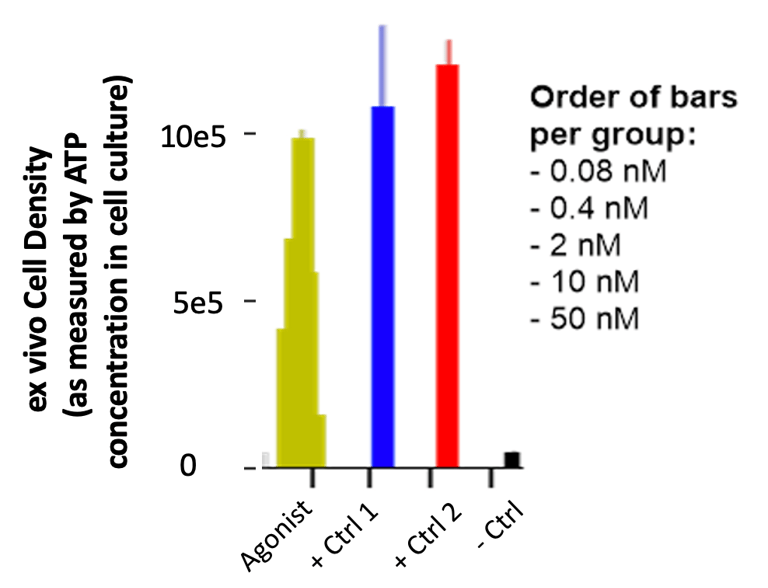
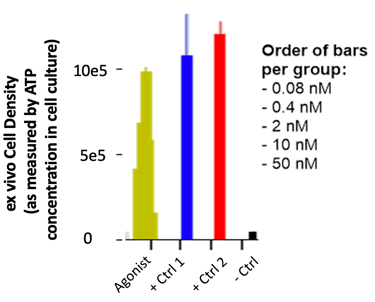
Receptor cross-linking is an important part of activation, so the agonist required high valency. The native ligands and a few commercial/clinical antibodies served as the basis for re-formatting the soluble agonist into mono-, bi-, tetra-, and hexaspecific formats. Some antibodies did much better than others given the same format; I think the location of the receptor epitope mattered so as to allow sufficient space for the agonist while maintaining sufficient receptor-proximity for the phosphorylation.
The data report expansion (via an ATP proxy) as a function of agonist concentration compared to two formats of the native ligand and a no-treatment control.
Yeast Surface-Display Platform
As a continuation of some graduate school work, I engineered a new form of yeast display. The advantage of this system was that it didn't rely on fusions to cell-wall proteins and so could display complex proteins like IgG. The platform biotinylated yeast, coated them in avidin, and then secreted antibodies fused to a biotinylation peptide. The biotinylated antibodies were captured on the surface of yeast where they could be displayed. Alternatively, the antibodies could be secreted directly from yeast if there was no avidin on the surface. An engineered secretory leader coupled with over-expression of a folding chaperone could get titers close to 1 mg/L. A schematic of the display system is shown below along with surface expression of a clinical IgG. I co-founded a company (Celexion) that used the platform to engineer proteins for clients. The three flow figures below show how typical rounds of enrichment look with improved ligand binding arising despite decreasing ligand concentration.
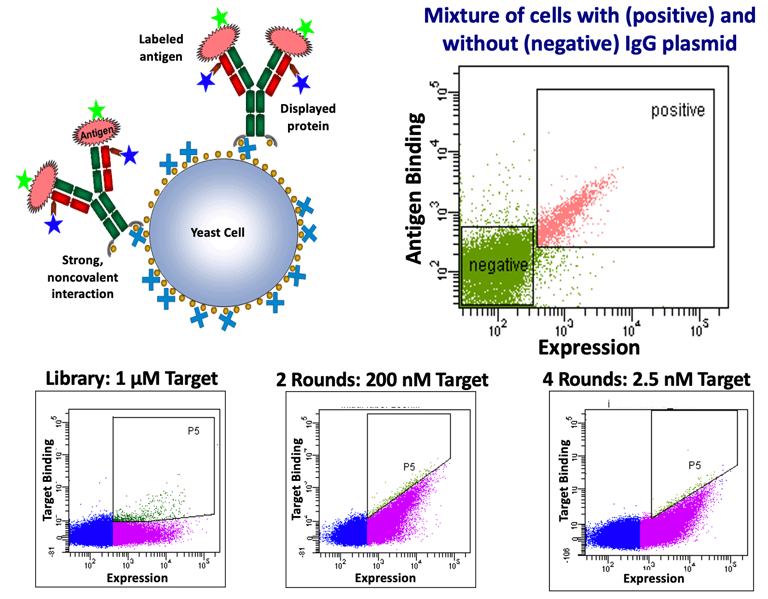
Naive IgG Antibody Library
To complement the display platform, we created a naive human antibody library. We used a semi-synthetic method that combined human germline V- and J-genes with B-cell derived CDR3s from IgM sequence.
We relied heavily on the Kabat database to distinguish contact residues from framework residues. We also biased the library toward V-genes that are most commonly expressed in humans.
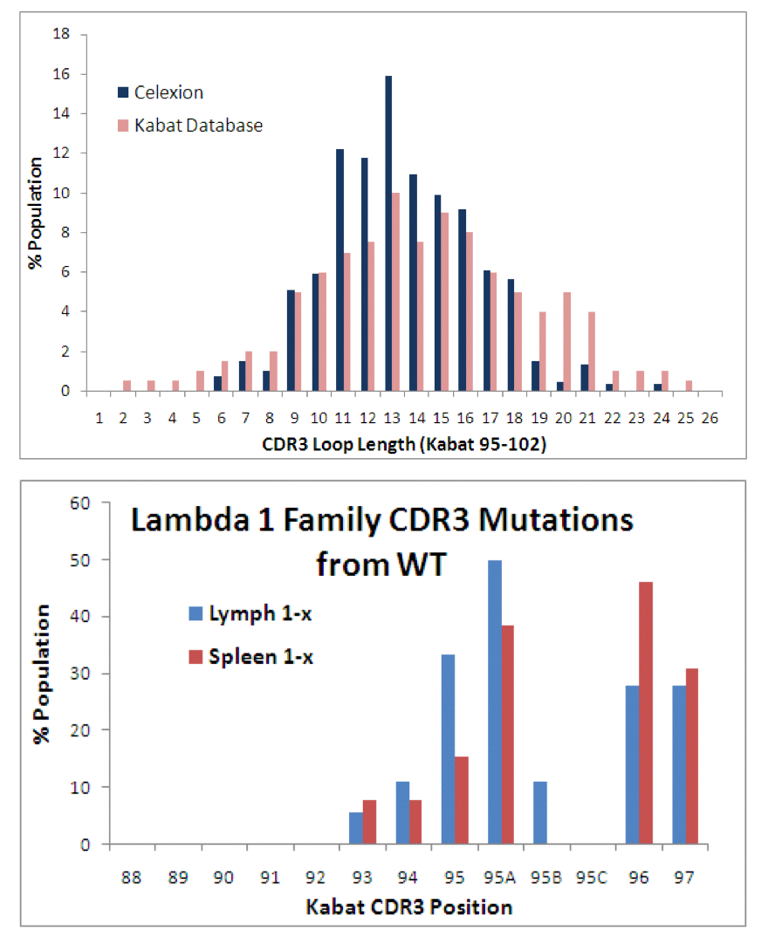
The heavy chain CDR3 length (at right) closely resembles the most comprehensive database of antibody sequence at the time (Kabat). We were also able to isolate diversity in the light-chain CDR3 (Lambda VL-1 family derived from lymph and spleenic B-cells shown at right). The library was laboriously transformed into yeast and approached 10 billion members.
Celexion used this library as a service as well as licensed the library and the display platform to partners. Please contact me if you would like to learn more about it.